The FoldamerDB contains information about foldamers collected from literature. It is designed to be intuitive and user-friendly, with tooltips and help icon. To get help for the specific part just click the
![]() which appears next to it. Clicking it will open a popup window with the help messages.
which appears next to it. Clicking it will open a popup window with the help messages.
FoldamerDB has options to browse all the foldamers, or browse by articles, or browse structures, or browse foldamers with biological activity. It also allows the user to perform a simple keyword search as well as chemical fingerprint based sub-structure search.
Nizami bilal, 2019.
Click here to view the paper: FoldamerDB paper
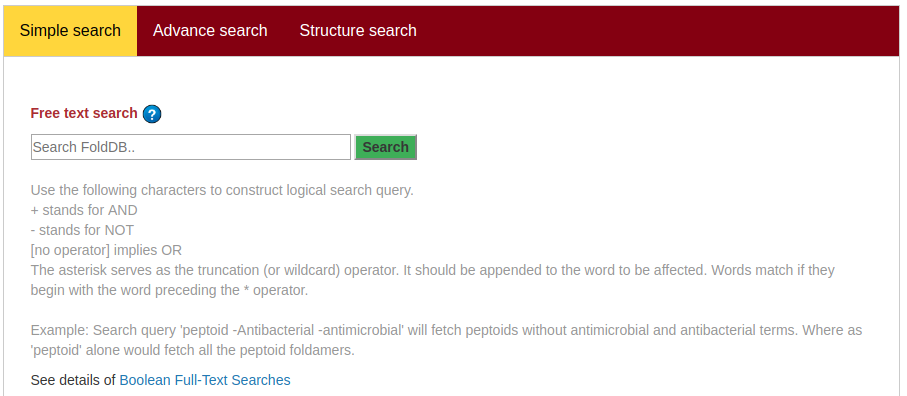
To perform free text search enter your query in the search box and click search. You can enter Sequence (one letter / three letter), Chemical name, Molecular formula, Application, Solvent, Type, and PDB ID. To search using specific criteria please go to advanced search or structure search tab.
Use the following characters to construct logical search query.
+ stands for AND
- stands for NOT
[no operator] implies OR
The asterisk serves as the truncation (or wildcard) operator. It should be appended to the word to be affected. Words match if they begin with the word preceding the * operator.
Example: Search query 'peptoid -Antibacterial -antimicrobial' will fetch peptoids without antimicrobial and antibacterial terms. Where as 'peptoid' alone would fetch all the peptoid foldamers.
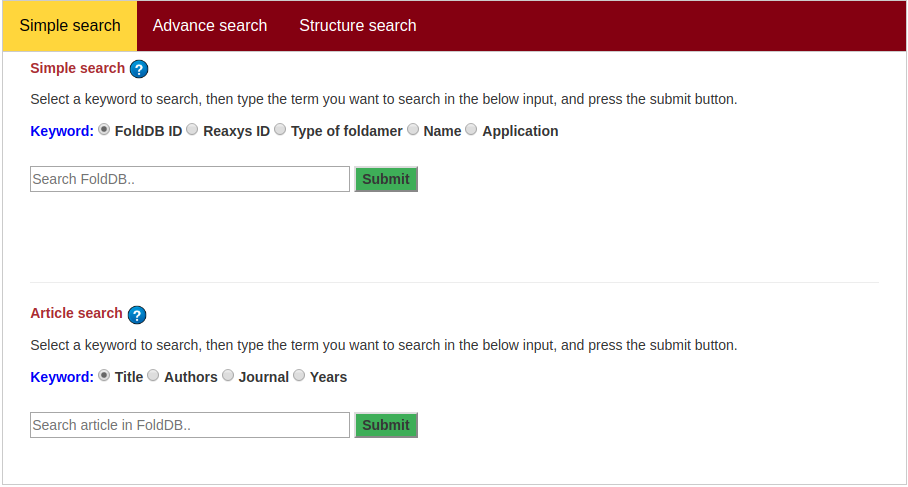
Select a keyword such as FoldDB ID, Application, Reaxys ID, and Type to search, then type the term you want to search in the given input, and press the submit button. To search in the articles, select a search field such as Title, Authors, Year, or Journal name, then type the term you want to search in the given input, and press the submit button.
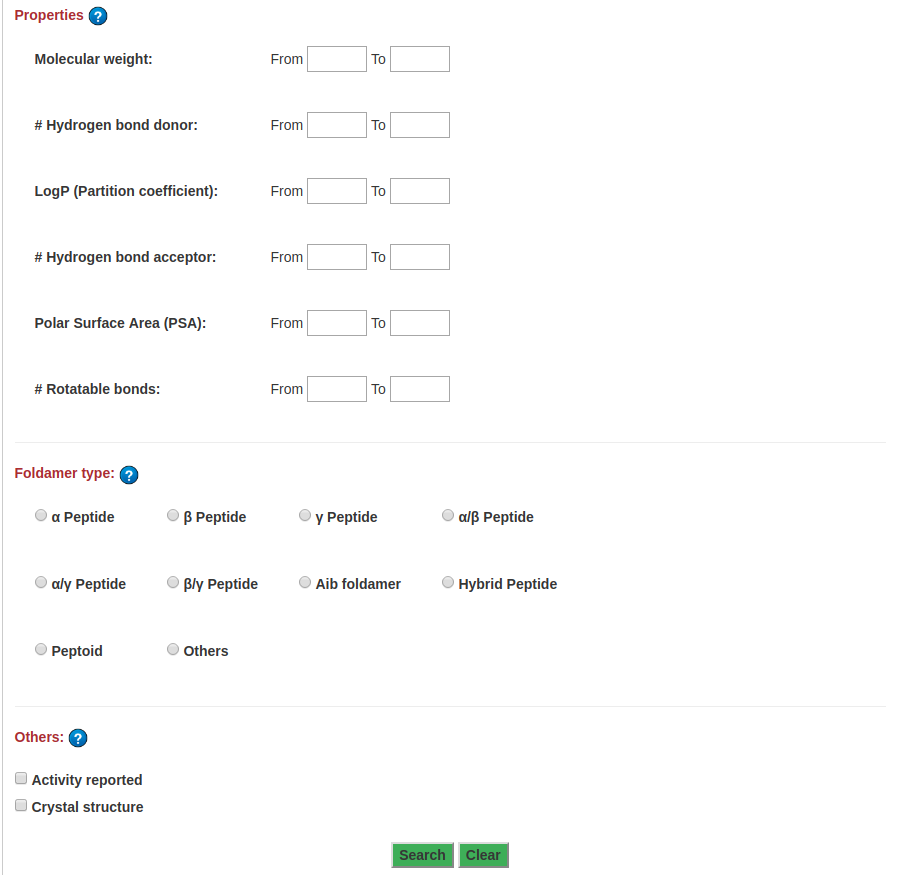
Advanced search allows to perform complex search based on multiple criteria. Fill upper/lower bound for one or more of the property values, select type of backbone, and check box under other category to perform the complex search. At least one field is required to search.
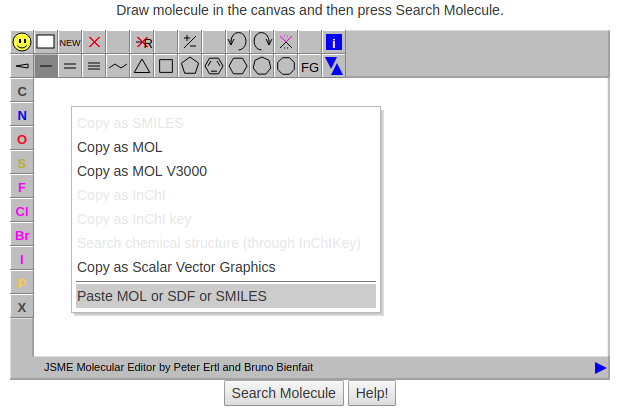
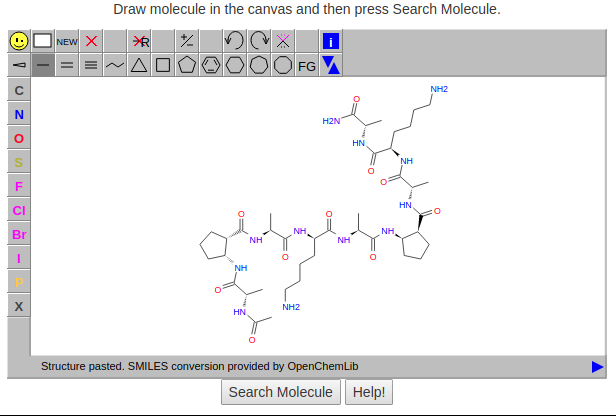
Sub-structure search allows to search foldamers by defining a chemical structure query. JSME Molecule Editor by Peter Ertl and Bruno Bienfait is used to supports drawing and editing of molecules. Users can draw a query molecule or can insert a valid SMILES string in the JSME canvas by right clicking (or click on  ) and then selecting 'Paste SDF or MOL or SMILES' . Once you have the query molecule in the canvas, press Search molecule to search. Click on the help button for more details (Figure 4 and Figure 5).
) and then selecting 'Paste SDF or MOL or SMILES' . Once you have the query molecule in the canvas, press Search molecule to search. Click on the help button for more details (Figure 4 and Figure 5).
The chemical similarity is measured by the help of chemicals fingerprints. The binary fingerprint consists of each molecule as an array of 32-bit hexadecimal numbers. Each key denotes the presence or absence of a particular substructure in a molecule. FoldamerDB uses openBabel [1] for calculating FP2 fingerprints (a path-based fingerprint which indexes small molecule fragments). 64 bit decimal form of the fingerpirnt is used with MySQL's binary arithmetic to search [2].
References
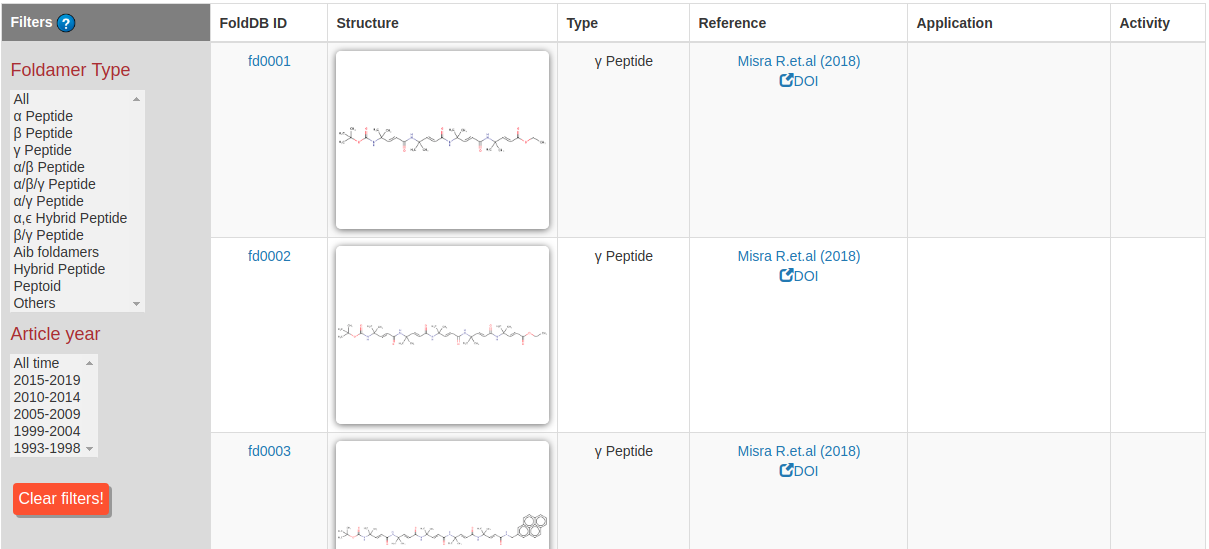
This page allow users to browse all the foldamers in a tabulated form. The table is spread over multiple pages, and a specific page can be accessed by clicking in the pagination menu.

The foldamer's table shows the structure of the foldamers, their FoldDB IDs, application, type, reference, and if the activity is reported or not, etc. The table can be filtered based on the type of backbone and year of publication. Clicking on a specific FoldDB ID opens the foldamer's entry page, where further details about it can be found (See FoldamerDB Foldamer Entry Section and Figure 10). Left mouse click on the structure to enlarge the image.
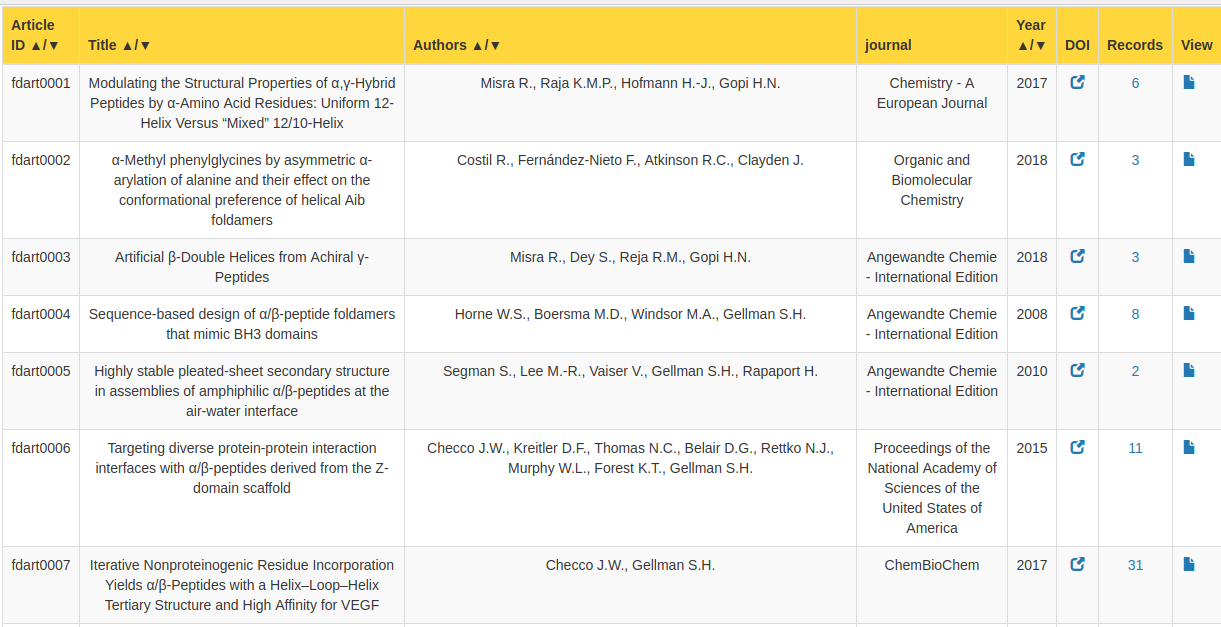
This page allow users to browse all the articles in the FoldamerDB in a tabulated form. The title of the article, author's name, year of publication, DOI number (open in new tab), Journal name and, number of records in the articles are shown. The table can be sorted by clicking on the column's heading with ▲/▼ symbol.
This page allow users to browse the foldamers with reported 3D crystal structure. Links are provided to the external databases such as Protein Data Bank (PDB)  and Cambridge Crystallography Database (CSD)
and Cambridge Crystallography Database (CSD)  .
.
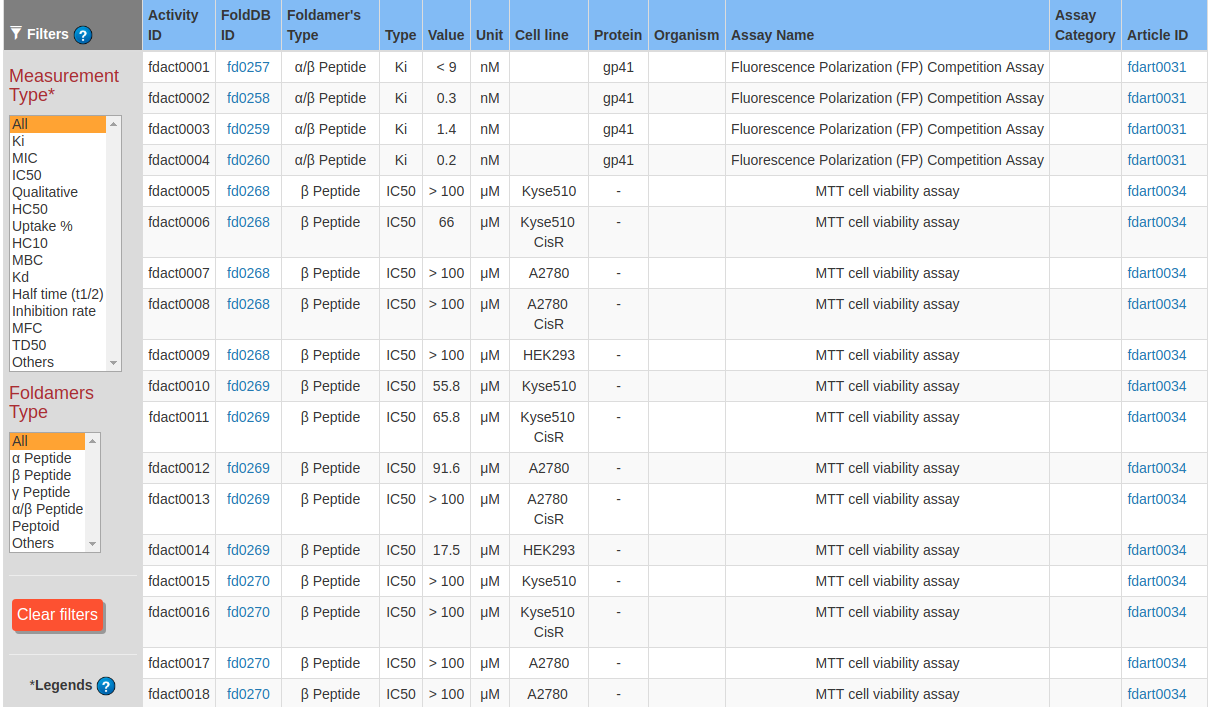
This page allow users to browse the biological activities of the foldamers. The activity table can be filtered using two criterias based on type of activity measurement and, backbone type. To select the type of measurement and backbone click on the corresponding filter on the left side of page.
This page allow users to access the detailed information on a single foldamer entry in FoldamerDB. The information in this page is devided into various sections. A molecular structure along with a 3D model of the foldamer is shown at the begining of each page. 3D model is rendered using Jsmol and various control buttons are provided to toggle the different molecular representations (Such as CPK, Ball&Stikc, and Stick etc.). To rotate the model click and hold the left button on the canvas area and move the mouse.
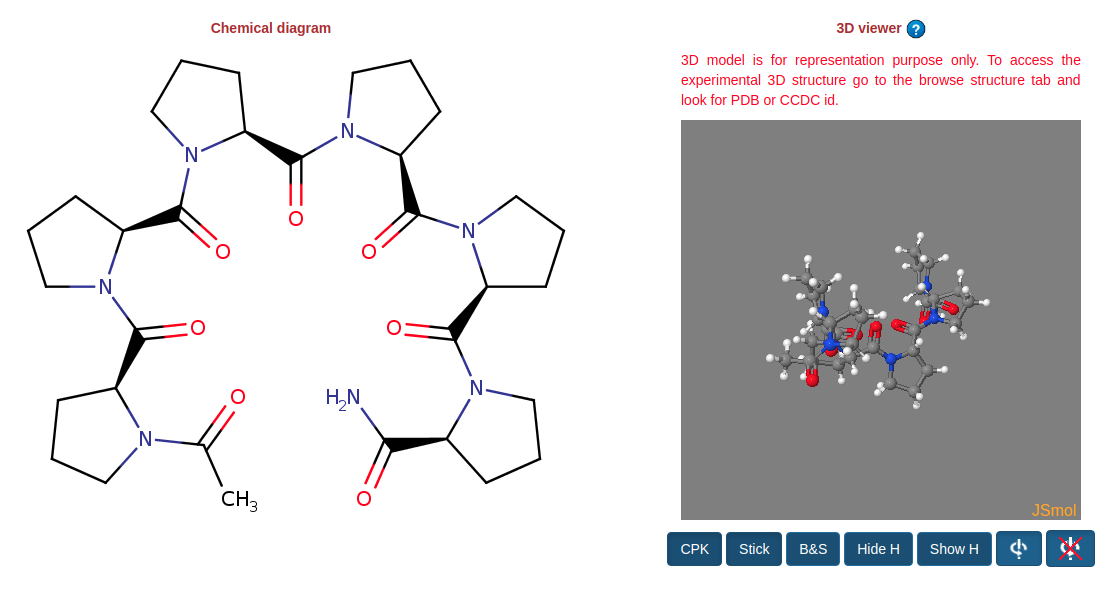
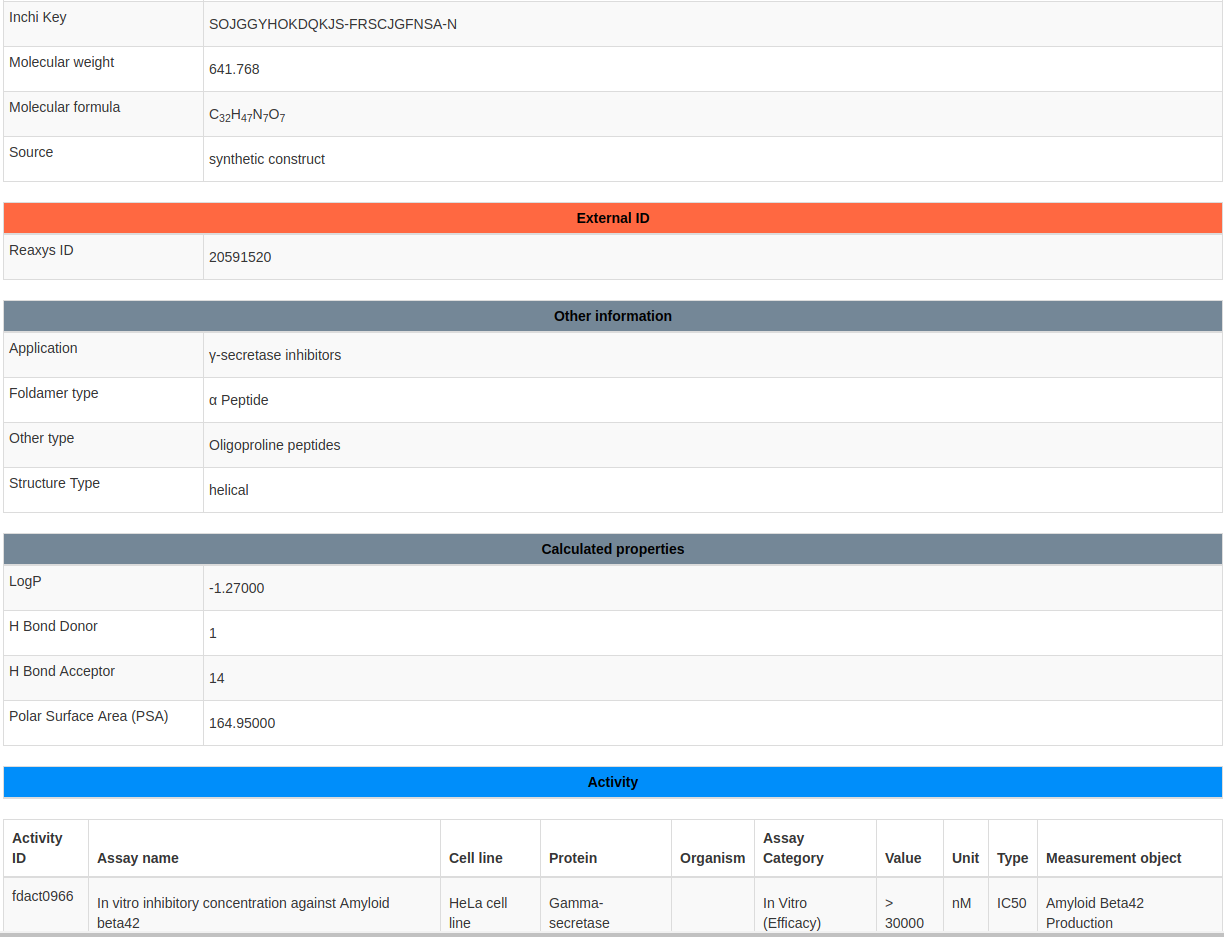
Next sections shows the information related to identifications, external IDs, structure, physico-chemical properties, biological activity, and bibliography of a single foldamers. Just below the activity block there is a link (Show all activity from this article ID:fdart0155) to browse all the activity reported in the single article ( see Figure 12 ).
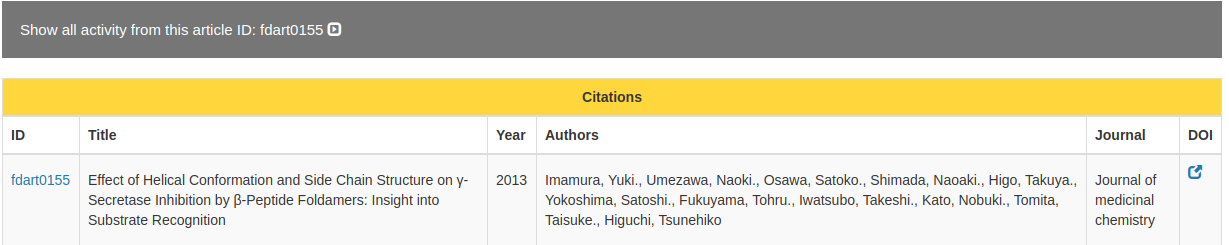
This page shows the details of a single article.
Title: e.g. 'Modulating the Structural Properties of α,γ-Hybrid Peptides by α-Amino Acid Residues: Uniform 12-Helix Versus “Mixed” 12/10-Helix'
Authors name: e.g. Misra R., Raja K.M.P., Hofmann H.-J., Gopi H.N.
Name of the Journal: e.g. Chemistry - A European Journal
Year of publication: e.g. 2017
FoldamerDB article ID: e.g. fdart0001
PUBMED ID: e.g. 28922503
DOI : Link to the extenral full text article.
Foldamers from this article can be browsed by clicking on .